"Twins" retrives similar metabolites in chemical structure.
|
|
"Twins" retrives similar metabolites in chemical structure. |
| last update : 2020.01.06 |
| INPUT WORD = C00027886 , 50% or more |
|
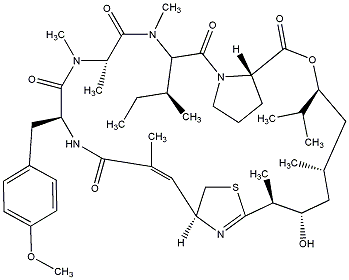
| [ Metabolite Name : Apratoxin C ] | |
| Number of matched data : 50 | |
| CID | Metabolite Name | Structural formula | Metabolite Activity | Similarity (%) |
|---|---|---|---|---|
| C00027885 | Apratoxin B | 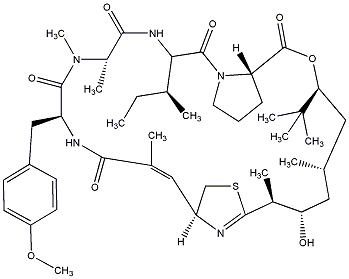 |
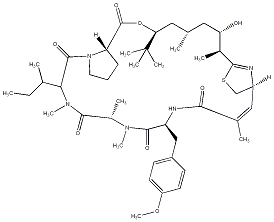
99.16 | C00034416 | Apratoxin A (-)-Apratoxin A |
 |
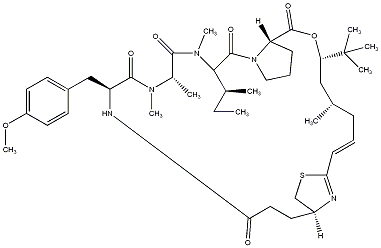
98.35 | C00038483 | Apratoxin E (-)-Apratoxin E |
 |
94.96 | C00038482 | Apratoxin D | 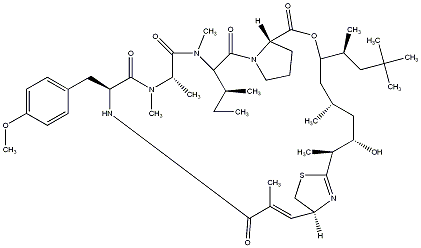 |
90.55 | C00034566 | Kulokekahilide 2 | 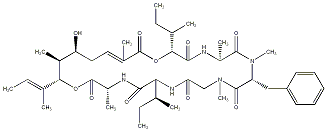 |
89.08 | C00034736 | Ulongapeptin | 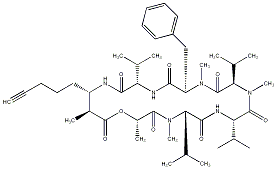 |
87.39 | C00039320 | Gypsin C | 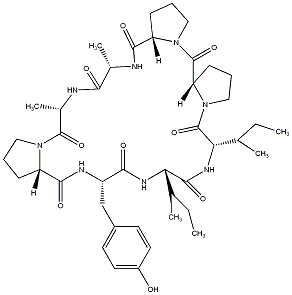 |
86.89 | C00028306 | Glabrin C | 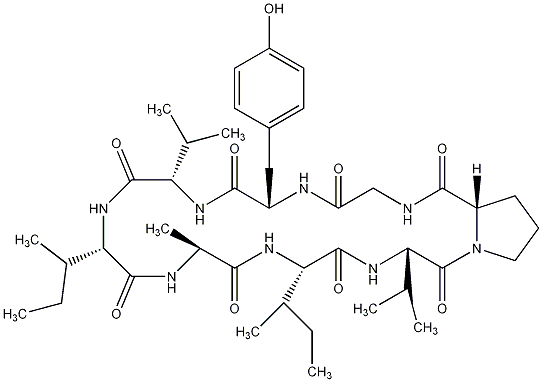 |
86.55 | C00028435 | Kulolide 1 | 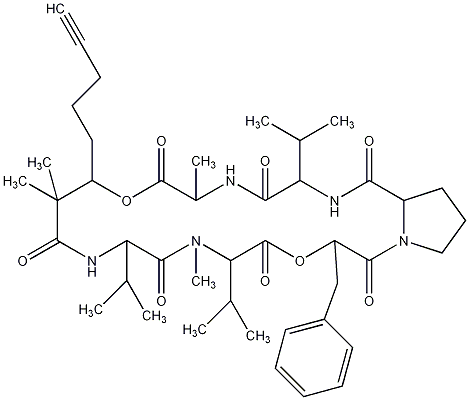 |
86.55 | C00028436 | Kulolide-2 | 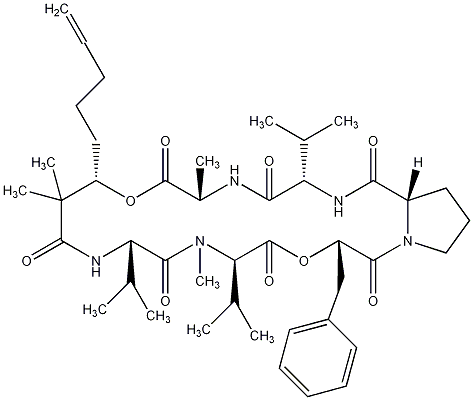 |
86.55 | C00028437 | Kulolide-3 | 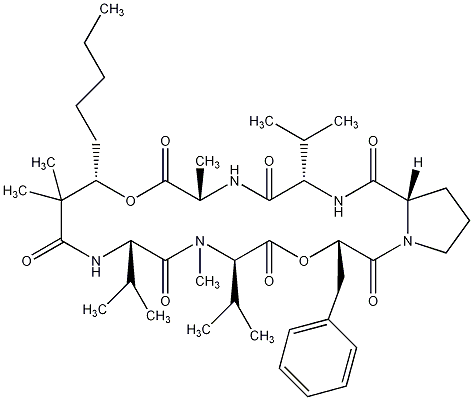 |
86.55 | C00034638 | Pitipeptolide A (-)-Pitipeptolide A |
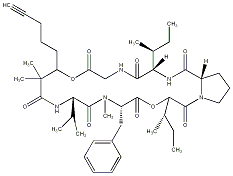 |
86.55 | C00034639 | Pitipeptolide B | 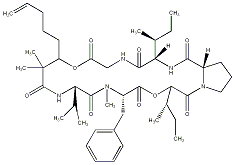 |
86.55 | C00015654 | Tubulysin B | 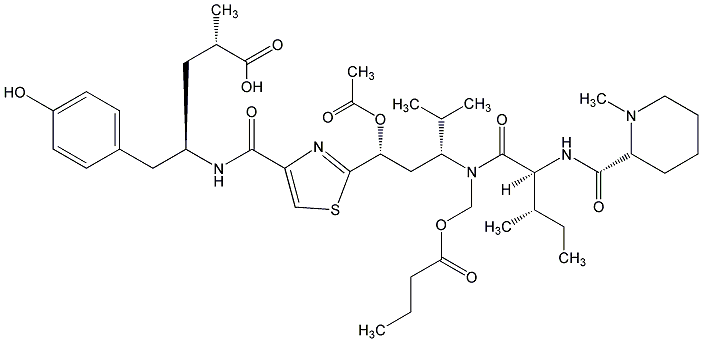 |
85.71 | C00028879 | Pseudostellarin F | 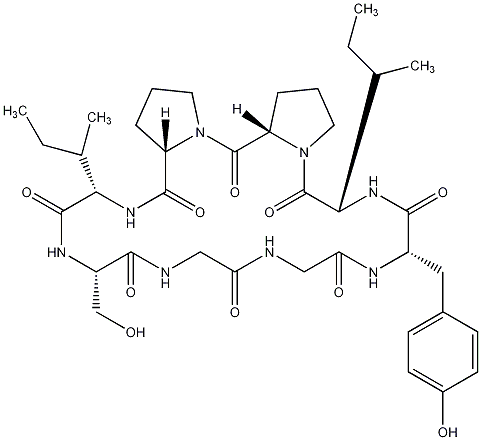 |
85.71 | C00018730 | I 337B | 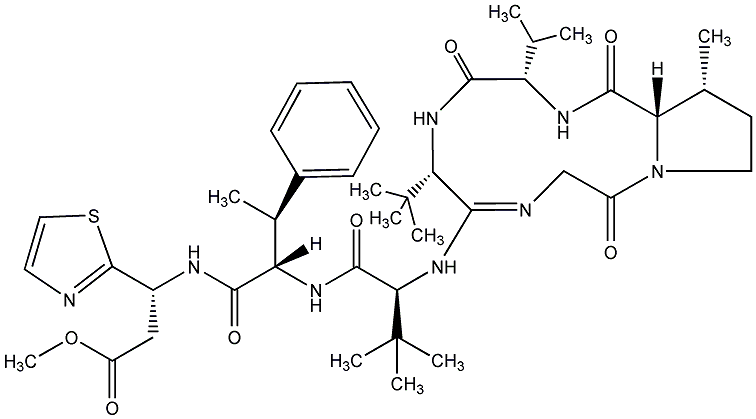 |
84.87 | C00046443 | Symplostatin 1 | 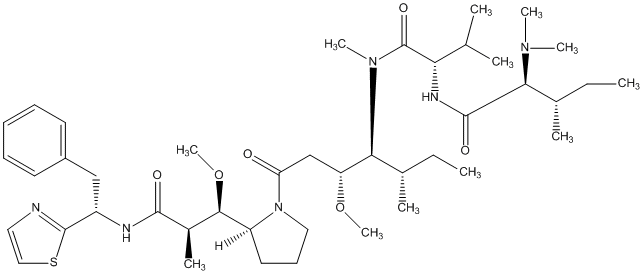 |
84.87 | C00048081 | Rolloamide A (-)-Rolloamide A |
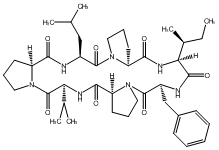 |
84.87 | C00034617 | Palau'amide | 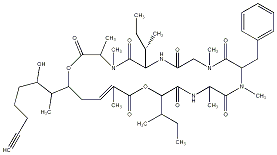 |
84.55 | C00015653 | Tubulysin A | 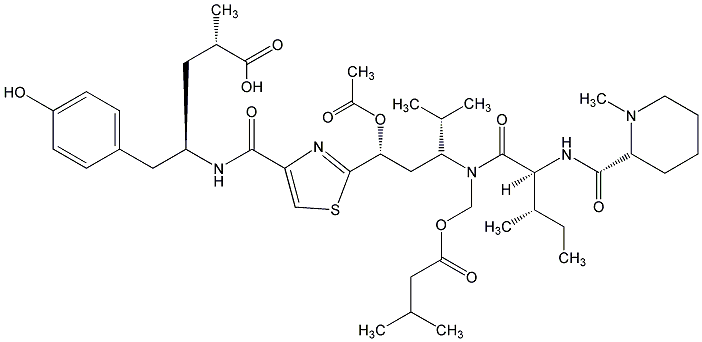 |
84.17 | C00015655 | Tubulysin D (+)-Tubulysin D |
 |
84.03 | C00015656 | Tubulysin E | 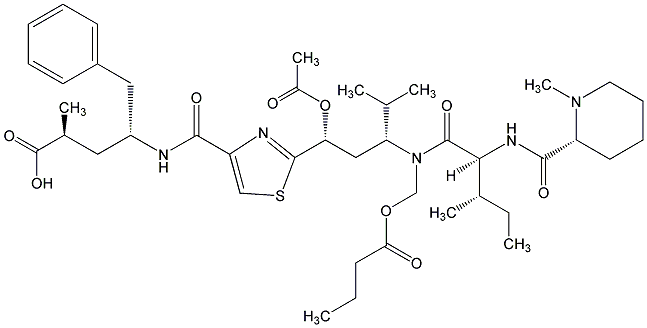 |
84.03 | C00044841 | Kahalalide E | 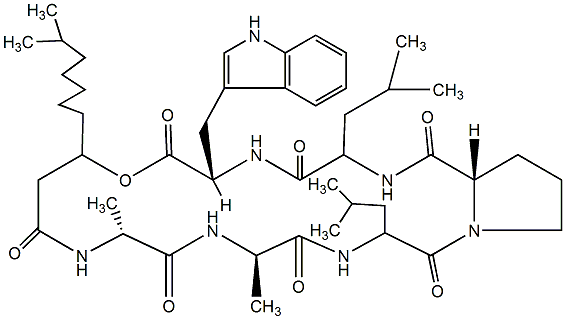 |
83.74 | C00041898 | Stylisin 1 (-)-Stylisin 1 |
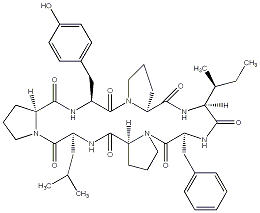 |
83.20 | C00028585 | Microginin 99A | 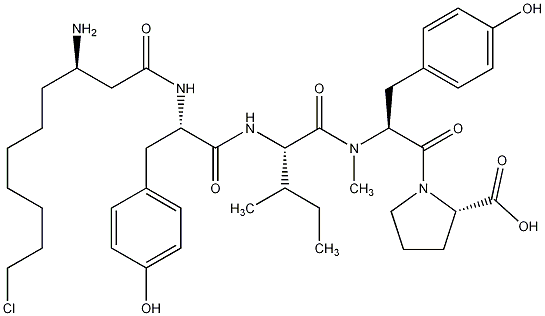 |
83.19 | C00029944 | Cherimolacyclopeptide G | 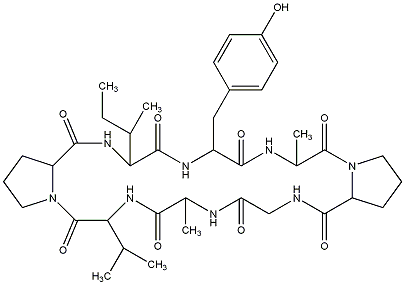 |
83.19 | C00040038 | Planktocyclin | 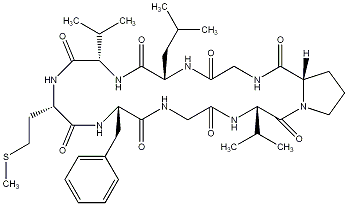 |
83.19 | C00047934 | Itralamide B | 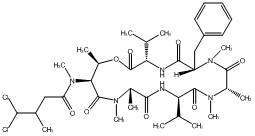 |
83.19 | C00048082 | Rolloamide B (-)-Rolloamide B |
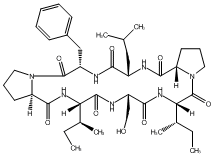 |
83.19 | C00041899 | Stylisin 2 | 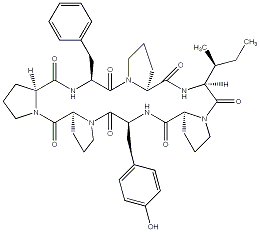 |
83.06 | C00047826 | Cycloaspeptide F (-)-Cycloaspeptide F |
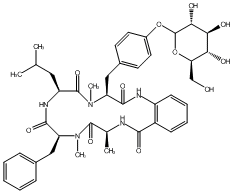 |
82.93 | C00044676 | Cyclotheonamide E4 | 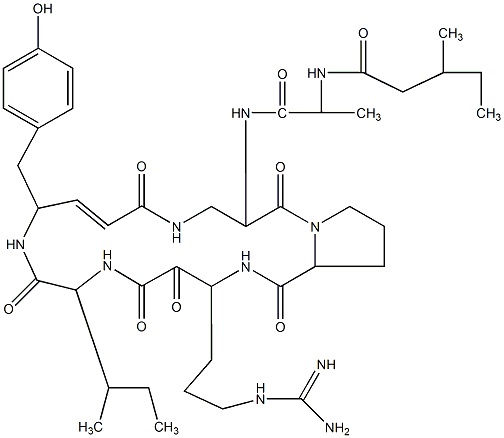 |
82.79 | C00027884 | Apramide G | 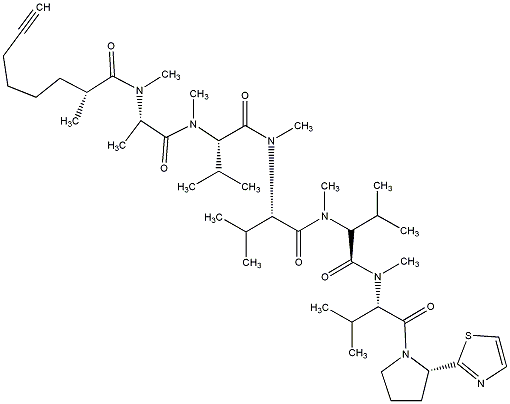 |
82.35 | C00028305 | Glabrin B | 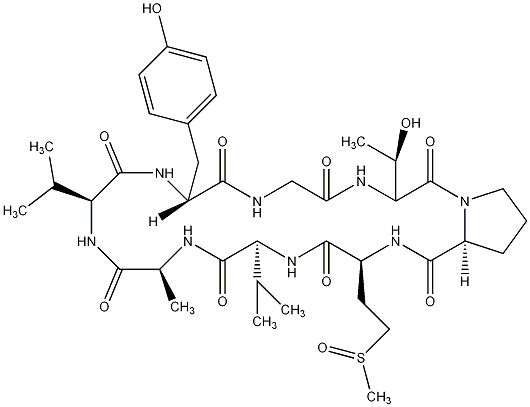 |
82.35 | C00028586 | Microginin 99B | 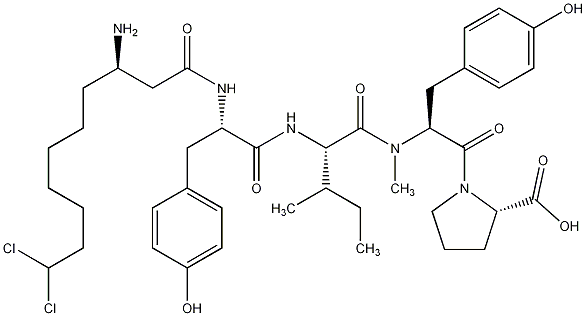 |
82.35 | C00031126 | RA IV | 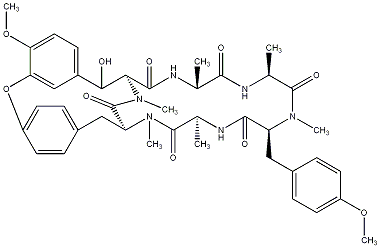 |
82.35 | C00031958 | Konbamide | 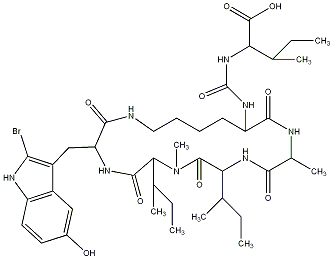 |
82.35 | C00046608 | Anabaenopeptin I (-)-Anabaenopeptin I |
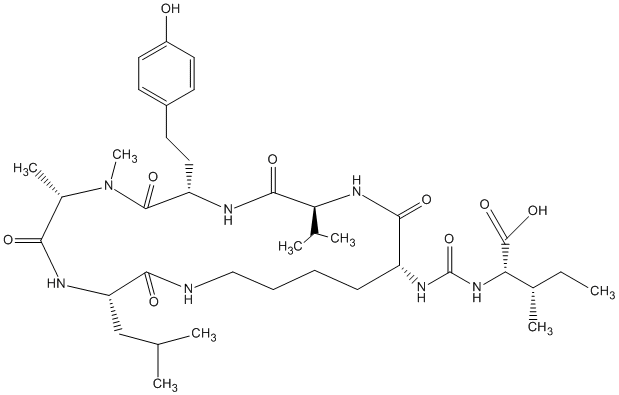 |
82.35 | C00049853 | Arenarin A | 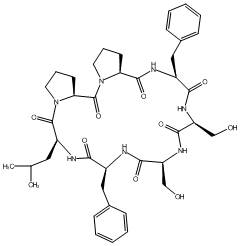 |
82.35 | C00028583 | Microginin 299C | 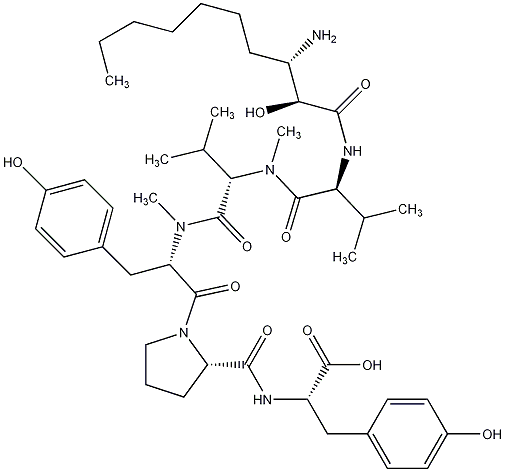 |
82.26 | C00041470 | Cycloleonuripeptide E | 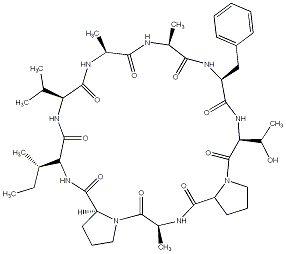 |
81.89 | C00024796 | Rhizonin A | 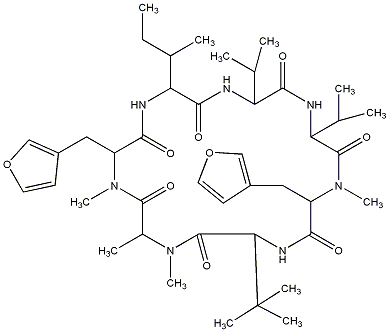 |
81.51 | C00028808 | Patellamide B | 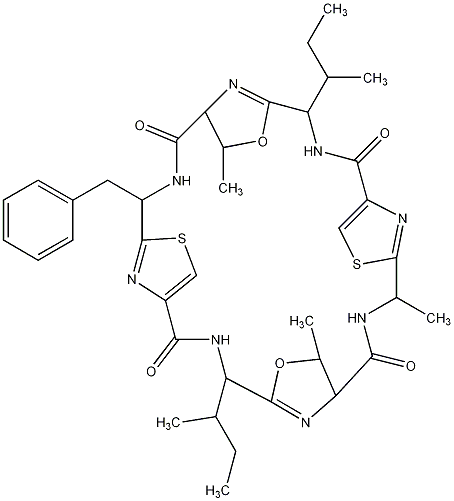 |
81.51 | C00028810 | Patellamide G | 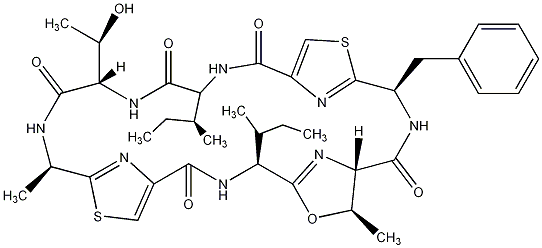 |
81.51 | C00034707 | Tasipeptin B | 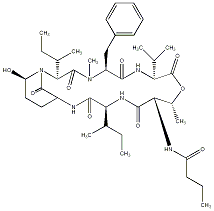 |
81.51 | C00041445 | Citrusin III | 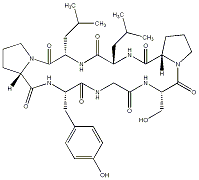 |
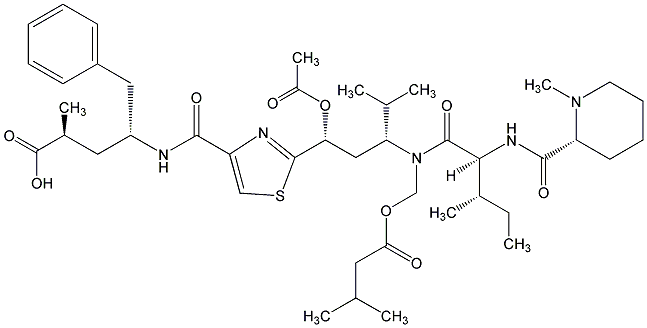
81.51 | C00044726 | Dolastatin 10 (-)-Dolastatin 10 |
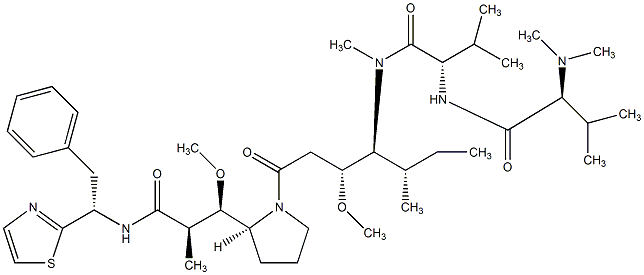 |
81.51 | C00045865 | CID is old! | 81.51 | C00028984 | Segetalin E | 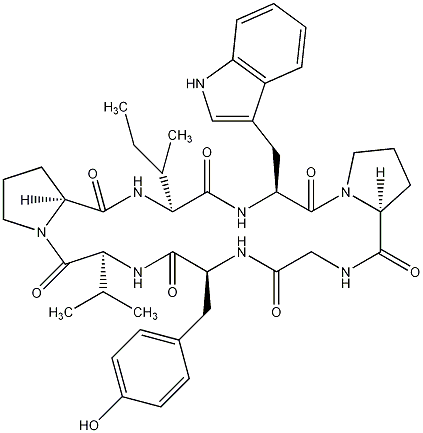 |
81.30 | C00028628 | Mycobactin J | 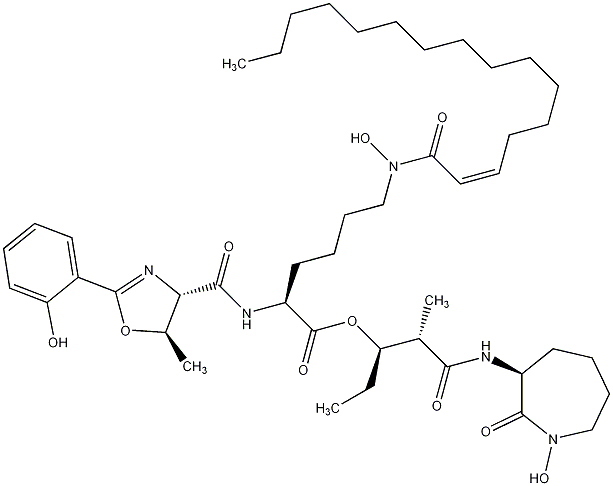 |
81.15 |